Zooplancton lagos crÁter de michoacan
Zooplancton epicontinental de los lagos cr├Īter de Michoac├Īn NOMBRE: Mar├Ła Guadalupe Ramos N├║├▒ez ESTATUS: Tesista de licenciatura ASESOR: M.C. Rub├®n Hern├Īndez Morales DEPENDENCIA: Laboratorio de Biolog├Ła Acu├Ītica ŌĆ£J. Javier Alvarado D├ŁazŌĆØ Facultad INSTITUCI├ōN: Universidad Michoacana de San Nicol├Īs de Hidalgo Descripci├│n El zooplancton es una comunidad biol├
 Biosci. Biotechnol. Biochem., 77 (4), 120936-1ŌĆō3, 2013
Highly E’¼ācient Transformation of the Diatom Phaeodactylum tricornutumby Multi-Pulse Electroporation
Mado MIYAHARA,1 Masaki AOI,1 Natsuko INOUE-KASHINO,2Yasuhiro K
1Graduate School of Biostudies, Kyoto University, Kyoto 606-8502, Japan2Graduate School and Faculty of Science, University of Hyogo, 3-2-1 Kohto, Ako-gun, Hyogo 678-1297, Japan
Received December 6, 2012; Accepted January 10, 2013; Online Publication, April 7, 2013
A highly e’¼ācient nuclear transformation method was
cultured in DaigoŌĆÖs IMK culture medium (Nihon
established for the pennate diatom Phaeodactylum
Pharmaceutical, Osaka, Japan) supplemented with sea
tricornutum using an electroporation system that drives
salts (Sigma, St. Louis, MO) and 0.2 mM Na2SiO3.
Biosci. Biotechnol. Biochem., 77 (4), 120936-1ŌĆō3, 2013
Highly E’¼ācient Transformation of the Diatom Phaeodactylum tricornutumby Multi-Pulse Electroporation
Mado MIYAHARA,1 Masaki AOI,1 Natsuko INOUE-KASHINO,2Yasuhiro K
1Graduate School of Biostudies, Kyoto University, Kyoto 606-8502, Japan2Graduate School and Faculty of Science, University of Hyogo, 3-2-1 Kohto, Ako-gun, Hyogo 678-1297, Japan
Received December 6, 2012; Accepted January 10, 2013; Online Publication, April 7, 2013
A highly e’¼ācient nuclear transformation method was
cultured in DaigoŌĆÖs IMK culture medium (Nihon
established for the pennate diatom Phaeodactylum
Pharmaceutical, Osaka, Japan) supplemented with sea
tricornutum using an electroporation system that drives
salts (Sigma, St. Louis, MO) and 0.2 mM Na2SiO3.


 Highly E’¼ācient Transformation of a Diatom by Electroporation
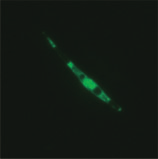
staining in the cells containing uidA (Fig. 3D), con’¼ürm-
ing that expression of the reporter genes was maintained
stably during the repeated segregation process. Expres-
sion level of sgfp in each transgenic line was estimatedfrom the intensity of GFP ’¼éuorescence (Fig. 3E). GFP
’¼éuorescence levels were di’¼Ćerent among the transgenic
cell lines and this was probably caused by di’¼Ćerences in
copy number or the position of the reporter gene in thegenome.
Highly E’¼ācient Transformation of a Diatom by Electroporation
staining in the cells containing uidA (Fig. 3D), con’¼ürm-
ing that expression of the reporter genes was maintained
stably during the repeated segregation process. Expres-
sion level of sgfp in each transgenic line was estimatedfrom the intensity of GFP ’¼éuorescence (Fig. 3E). GFP
’¼éuorescence levels were di’¼Ćerent among the transgenic
cell lines and this was probably caused by di’¼Ćerences in
copy number or the position of the reporter gene in thegenome.